Comparative Evaluation of Genome Assemblers from Long-Read Sequencing for Plants and Crops
植物和作物基因组组合的比较评价
- Hyungtaek Jung*, Min-Seung Jeon, Matthew Hodgett, Peter Waterhouse, and Seong-il Eyun*
Abstract
The availability of recent state-of-the-art long-read sequencing technologies has significantly increased the ease and speed of producing high-quality plant genome assemblies. A wide variety of genome-related software tools are now available and they are typically benchmarked using microbial or model eukaryotic genomes such as Arabidopsis and rice. However, many plant species have much larger and more complex genomes than these, and the choice of tools, parameters, and/or strategies that can be used is not always obvious. Thus, we have compared the metrics of assemblies generated by various pipelines to discuss how assembly quality can be affected by two different assembly strategies. First, we focused on optimizing read preprocessing and assembler variables using eight different de novo assemblers on five different Pacific Biosciences long-read datasets of diploid and tetraploid species. Then, we examined a single scaffolding tool (quickmerge) that has been employed for the postprocessing step. We then merged the outputs from multiple assemblies to produce a higher quality consensus assembly. Then, we benchmarked the assemblies for completeness and accuracy (assembly metrics and BUSCO), computer memory, and CPU times. Two lightweight assemblers, Miniasm/Minimap/Racon and WTDBG, were deemed good for novice users because they involved smaller required learning curves and light computational resources. However, two heavyweight tools, CANU and Flye, should be the first choice when the goal is to achieve accurate and complete assemblies. Our results will provide valuable guidance in future plant genome projects and beyond.
最近最先进的长读测序技术的可用性极大地提高了生产高质量植物基因组组装的便捷性和速度。
现在有各种各样与基因组相关的软件工具,它们通常使用微生物或真核生物基因组模型(如拟南芥和水稻)作为基准。
然而,许多植物物种的基因组要比这些大得多、复杂得多,而可使用的工具、参数和/或策略的选择并不总是显而易见的。
因此,我们比较了各种管道生成的装配指标,以讨论两种不同的装配策略如何影响装配质量。
首先,我们专注于优化读取预处理和组装变量,使用八种不同的从头组装程序,对太平洋生物科学公司的五种不同的二倍体和四倍体物种的长期读取数据集。
然后,我们一个脚手架检查工具(quickmerge)被用于后处理步骤。
然后,我们合并来自多个组件的输出,以产生更高质量的一致组件。
然后,我们对组装的完整性和准确性(组装指标和BUSCO)、计算机内存和CPU时间进行基准测试。
两种轻量级的组装程序Miniasm/Minimap/Racon和WTDBG被认为是适合新手用户的,因为它们需要更小的学习曲线和更少的计算资源。
然而,当目标是实现精确和完整的装配时,两种重量级工具CANU和Flye应该是首选。
我们的研究结果将为未来的植物基因组计划提供有价值的指导。
KEYWORDS:
- plant genome
- next-generation sequencing
- Pacific Biosciences
- long reads
- nanopore
- assemblers
Introduction
Introduction
ARTICLE SECTIONS
Jump To
The advent of next-generation sequencing (NGS) technologies has initiated a new era in genomics research. Despite dramatic improvements in DNA sequencing technologies and computation tools, assemblies using short reads remain very challenging because of large quantities of repetitive content in large genomes, uneven sequencing coverage, and the presence of (nonuniform) sequencing errors and chimeric reads.(1?3) To overcome the issues of NGS, long-read sequencing (LRS) technologies (Pacific Biosciences [PacBio] and Oxford Nanopore Technology) have been developed and recently actively adopted by the plant genomics community.(3?9) While emerging LRSs offer very long true reads (up to 200 kb in PacBio and 2 Mb in Nanopore), these technologies are still expensive (cost per base) and subject to high sequencing error rates (5–10% for PacBio and 10–15% for nanopore)(3,10) compared to NGS. However, applications of LRS to plant genomes have obvious advantages that are provided by long reads in de novo assembly, such as higher contiguity, smaller gaps, and fewer errors.(4,6,8,11?13) Nonetheless, care is required when planning a genome project to maximize assembly quality, cost, extra subchromosome length scaffolding (e.g., BioNano and Hi-C), and choice of assemblers.(3)
Determining which assembler can be used to produce the best quality assembly requires particular attention. The appropriate choice could depend on the size and complexity (repeat content, ploidy, etc.) of the genome to be assembled and the type of sequencing technology used to produce the input reads (e.g., NGSs vs LRSs).(3) While comparative evaluations such as the genome assembly gold-standard evaluation(14) and Assemblathon(15) can provide general guidelines, there is currently no systematic way to determine which assembler and parameter settings would produce the best assembly for a specific genome and/or dataset. Consequently, it is common practice to generate multiple genome assemblies from a few different assemblers, parameters, and algorithms [e.g., de Bruijn Graph (DBG) and overlap-layout-consensus (OLC)]. Then, researchers attempt to predict the best assembly based on assembly statistics, spot-checking, homology analysis, agreement with physical/genetic maps, and so forth. However, what constitutes the best assembly remains undefined. Because no perfect and error-free assembly exists, we must decide whether it is more important to maximize contig and scaffold length or minimize the number of misassemblies.(16,17)
To make matters more difficult, these new technologies and algorithms for long reads are typically benchmarked on microbial genomes or, if they are scaled appropriately, the human genome.(3,18) Unfortunately, the human genome is not the representative of all eukaryotic genomes; in particular, plant genomes are larger and more repetitive than human genomes, and plant biology (e.g., chloroplasts and mitochondrial genomes) makes obtaining high-quality DNA free from contaminants difficult.(3,18) Thus, technologies that work well on vertebrate genomes may not work well with plant genomes(19) because each assembler implements slightly different heuristics to deal with repetitions, uneven coverage, sequencing errors, and chimeric reads while assembling genomes. Furthermore, each sequencing platform comes with its own input and computational requirements, qualities of output and, naturally, labor and material costs. Nonetheless, the final assembly is very rarely finished to feature one solid sequence per chromosome. Instead, typical outputs are presented as unordered/unoriented sets of contiguous regions called contigs. Alternatively, assembly reconciliation (or postprocessing) algorithms have been created to both produce a higher quality consensus assembly by merging two or more draft assemblies and enhance the contiguity of the resultant assembly while avoiding introducing assembly errors.(17)
In this paper, we compare eight different long-read de novo genome assemblers (termed preprocessing here) for five different plant PacBio RS II reads and one reconciliation assembler (termed postprocessing here). The genome of the targeted species varies in size from 0.4 to 2.3 Gb and comprises diploids and allotetraploids. Using the PacBio long reads, we carried out a comprehensive evaluation of assemblers by measuring the quality of the consensus assembly. Our results can be used as an initial guide for further sequencing assembly projects and a basis for plant genome studies, as each assembly method has its own biases.
新一代测序技术的出现开启了基因组学研究的新时代。
尽管DNA测序技术和计算工具有了巨大的进步,但由于大基因组中存在大量重复内容、测序覆盖不均、测序错误(不均匀)和嵌合读数的存在,使用短读的组装仍然非常具有挑战性。
读测序(LRS)技术(太平洋生物科学(PacBio)和牛津纳米孔技术)最近开发并积极采用植物基因组学社区。(3?9)尽管新兴LRS提供很长的真读(200 kb PacBio和纳米孔的2 Mb),这些技术仍然是昂贵的(成本/基地)和受高测序错误率(PacBio 5 - 10%, 10 - 15%纳米孔)(10)门店。
然而,LRS植物基因组的应用有明显的优势,提供长途读入新创组装,如更高的接触,更小的差距,和更少的错误。(4、6、8、11?13)尽管如此,护理时需要计划一个基因组项目最大化装配质量、成本、额外subchromosome长度脚手架(例如,BioNano和高c),和汇编器的选择。(3)
决定哪个装配商可以用来生产最好质量的装配需要特别注意。
适当的选择可能取决于大小和复杂性(重复内容、倍性等)的基因组组装和测序技术用于生产的类型输入读取(例如,总会在vs lrs)。(3)而比较评估,如基因组组装为对照评价(14)和Assemblathon(15)可以提供一般指导方针,
目前还没有系统的方法来确定哪个装配程序和参数设置会为特定的基因组和/或数据集产生最好的装配。
因此,从一些不同的装配器、参数和算法中生成多个基因组装配是常见的做法。
, de Bruijn图(DBG)和重叠-布图共识(OLC)]。
然后,研究人员试图根据装配统计、抽查、同源性分析、与物理/遗传图谱的一致性等来预测最佳装配。
但是,什么是最好的程序集仍然没有定义。
由于不存在完美和无错误的组装,我们必须决定是将contig和支架的长度最大化,还是将错误组装的数量最小化更重要。
让事情变得更困难的是,这些用于长时间读取的新技术和算法通常以微生物基因组为基准,或者,如果它们被适当地缩放,以人类基因组为基准(3,18)。不幸的是,人类基因组不是所有真核生物基因组的代表;
特别是,植物基因组大,比人类基因组重复,和植物生物学(例如,叶绿体和线粒体基因组)使得获取高质量的DNA免受污染物困难。(3、18)因此,技术工作在脊椎动物基因组可能不适合与植物基因组(19)因为每个汇编程序实现了稍微不同的启发式处理重复,不均匀覆盖,测序错误,嵌合组装基因组时读取。
此外,每个测序平台都有自己的输入和计算要求、输出质量,当然还有人工和材料成本。
尽管如此,最后的组装很少完成,每个染色体都有一个固定的序列。
相反,典型的输出被表示为无序/无定向的连续区域集合,称为contigs。
另外,装配协调(或后处理)算法已经被创建,通过合并两个或更多的草图装配产生更高质量的一致装配,并增强结果装配的连续性,同时避免引入装配错误。
在本文中,我们比较了八种不同的长读从头基因组装配器(这里称为预处理)对五种不同的植物PacBio RS II读取和一个调节装配器(这里称为后处理)。
目标物种的基因组大小从0.4到2.3 Gb不等,包括二倍体和异源四倍体。
使用PacBio long reads,我们通过测量consensus组装的质量对组装者进行了综合评价。
我们的研究结果可以作为进一步测序装配项目的初始指导,也可以作为植物基因组研究的基础,因为每种装配方法都有其各自的偏倚。
Materials and Methods
ARTICLE SECTIONS
Jump To
Input Sequence and Species
Five plant genomes were selected to evaluate the performance of multiple genome assemblers because their PacBio RSII reads had more than 50X coverage for each genome and were available at the National Center for Biotechnology Information (NCBI) (Table 1). See the previously published papers for more library preparation and sequencing information (Table 1).
Table 1. Summary of Input Sequence and Species
| species category | Arabis alpina | O. indica | Durio zibethinus | C. quinoa | Z. mays B73 |
|---|---|---|---|---|---|
| sequencing platform | RSII | RSII | RSII | RSII | RSII |
| genome size (Gb) | 0.37 | 0.40 | 0.74 | 1.40 | 2.30 |
| ploidy | diploid | diploid | diploid | allotetraploid | diploid |
| coverage (X) | 86 | 118 | 153 | 54 | 65 |
| accession numbers | ERX1795357, PRJNA241291 | PRJCA000313 | PRJNA400310 | PRJNA306026 | SRX1472849 |
| references | Jiao et al. 2017(5) | Du et al. 2017 | Teh et al. 2017 | Jarvis et al. 2017 | Jiao et al. 2017(9) |
Assembly and Evaluation
Eight emerging de novo assemblers and pipelines (preprocessing assembly) were tested on a cluster-based high-performance computer (HPC) using PBSpro for job scheduling and workload management at the Queensland University of Technology in Australia and Chung-Ang University in Korea. Because of the specific requirements in computing environments, only five assemblers (with default parameters) were further compared in terms of assembly performance: CANU (ver. 1.7),(20) Flye (ver. 2.3.6),(21) Miniasm (r129)/Minimap/Racon (MMR),(22,23) SMARTdenovo (SMD) (https://github.com/ruanjue/smartdenovo), and WTDBG (ver. 1.2.8) (https://github.com/ruanjue/wtdbg). Three unsuccessful assemblers (Falcon, Hinge, and MECAT) were not included for further comparison (Figure 1). To ensure fairness in the comparisons, the following two criteria were applied to this work: (1) if an error correction and a polishing stage were embedded in the assembler and the pipeline, we processed it until its final assembly output and (2) further polishing such as PacBio (Arrow and Quiver) and/or Illumina (Pilon) was not considered for the final assembly output. However, Racon (mapping with minimap) was used as secondary error correction and consensus calling for the final contigs of WTDBG, and an embedded pipeline was used for MMR.(23) As a postprocessing step in the assembly process, we also employed quickmerge (QMG), a simple, fast, and general meta-assembler that merges assemblies to generate a more contiguous assembly.(24)
Figure 1
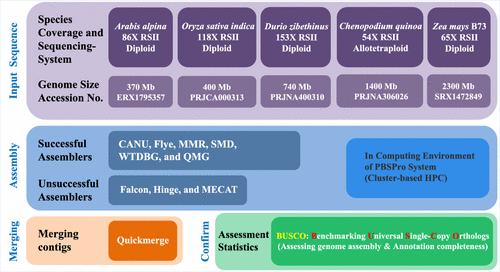
Figure 1. Summary of long-read assembly workflow and evaluation.
For completeness and contiguity, we used the Benchmarking Universal Single-Copy Orthologs (BUSCO) core plant dataset (ver. 3.0.2 and lineage db: embryophyta_odb9) to evaluate the gene contents(25) and calculated a full range of metrics for each assembly (such as a statistical summary of N50 length). To measure the computing time, we used the CPU time and memory usage (MEM) if the assembler could be done in a single-node job (24 cores with 240 Gb RAM or 192 cores with 6 Tb RAM); we used the actual wall-time and total memory usage for CANU if it required a multibranched job (multithreads and nodes) (Table 2).
Table 2. Summary of Computing Resources
| A. alpina | O. indica | D. zibethinus | C. quinoa | Z. mays | ||
|---|---|---|---|---|---|---|
| CANU | wall timea | 45 | 47 | 95 | 167 | 288 |
| MEM usageb | 1440 | 1440 | 1440 | 1440 | 1440 | |
| Flye | CPU timea | 626.53 | 192.57 | 1037.52 | 1708.46 | 6037 |
| MEM usageb | 303 | 136 | 465 | 395 | 1260 | |
| MMR | CPU timea | 252.03 | 36.32 | 523.42 | 1030.65 | 3224.30 |
| MEM usageb | 207 | 52 | 215 | 445 | 654 | |
| SMD | CPU timea | 392.45 | 75.32 | 358.55 | 78.28 | 2421 |
| MEM usageb | 40 | 20 | 45 | 38 | 962 | |
| WTDBG | CPU timea | 24.41 | 47.22 | 37.47 | 78.22 | 393.59 |
| MEM usageb | 45 | 103 | 49 | 93 | 315 | |
| QMG | CPU timea | 1 | 4 | 5 | 13 | 47 |
| MEM usageb | 55 | 67 | 118 | 175 | 376 |
a All times are in hours. b All memory usage (MEM) indicate the maximum memory usage (Gb).
Results
ARTICLE SECTIONS
Jump To
The results of this study are presented in two parts. First, we compared the five successful long read-based assemblers as the preprocessing portion. Second, we merged all assembled contigs in QMG as the postprocessing step. By validating the assemblies for sequence accuracy, we found both strengths and weaknesses and found that different methods resulted in stark differences in DNA sequence complexity, time, computational requirements, and cost.
Preprocessing Assembly Performance
The first stage of an assembly is to piece together long reads to form long contigs. Here, we focus on assessing the various existing pipelines and assemblers and comparing the results obtained from PacBio RSII data. While the long reads provided by PacBio can be used to generate a de novo assembly either alone or in conjunction with Illumina data, we show examples of assemblies from pure long reads.
We selected five successful assembly pipelines for LRS data (Figures 1 and 2). CANU, MMR, SMD, and WTDBG are based on the OLC algorithm. Flye is based on a generalized DBG algorithm. While CANU includes a base-error correction step, other assemblers do not require reads that have been error-corrected. After an initial assessment of the BUSCO scores, Miniasm/Minimap and WTDBG showed relatively low values (Figure 3). Thus, a third-party program called Racon was used as secondary error correction and consensus calling for the final contigs of these two assemblers because it aligns raw long reads to the contigs and generates a consensus, thus significantly increasing the initial accuracy.(23) Details on statistics and BUSCO on these assemblers and pipelines can be found in Figures 2 and 3. While no single assembler outperforms all others across all species, the highest assembly quality was observed in CANU/Flye followed by MMR. SMD and WTDBG showed the lowest accuracy in our attempts; as such, they might require a substantial polishing step, as they have no built-in error-correction stage.
Figure 2

Figure 2. Summary of assembly statistics and metrics. Aa: Arabis alpina; Os: O. indica; Dz: Durio zibethinus; Cq: C. quinoa; Zm: Z. mays B73. Note that three contigs are longer than 10 Mb in Cq QMG (E). (A) Total number of contigs; (B) total assembled contig size (Mb); (C) longest contig (Mb); (D) number of contigs > 100 kb; (E) number of contigs > 1 Mb; and (F) Nb50 of contigs (Mb).
Figure 3

Figure 3. Assessment of genome assembly and annotation completeness with BUSCO. (A) A. alpina; (B) O. indica; (C) Durio zibethinus; (D) C. quinoa; and (E) Z. mays B73. BUSCO scores indicate the relative levels of completeness and putative gene duplications (see Materials and Methods). In BUSCO graphs, the X-axis represents BUSCO scores (%), and the Y-axis represents assemblers (CANU, Fly, MMR, SMD, WTDBG, and QMG). The different colors represent the following: dark-blue square: complete (C) and duplicated (D). Light-blue square: complete (C) and single-copy (S). Yellow square: fragmented (F). Red square: missing (M).
Given the computing environment (an HPC with job scheduling and workload management) and dataset, all tested assemblers were relatively user-friendly. Flye, SMD, and WTDBG required fewer than three script lines/commands, but CANU and MMR needed ?20 script lines/commands. All executed scripts are summarized and available in the Supporting Information. Differences were observed in contiguity, completeness, and computing resources (CPU time, wall time, and memory usage). For Flye, if a target species was a large and complex genome (>2 Gb; Zea mays), it required specification in the subset of reads for initial contig assembly (e.g., --asm-coverage 30) because of memory and/or performance optimizations as opposed to the full input of reads. Despite the size and complexity of genomes, CANU and Flye showed reliable contiguity and completeness; however, the results provided by MMR, SMD, and WTDBG fluctuated (Figures 2 and 3). CANU required the most computing power (with larger memory and longer running time) followed by Flye, MMR, SMD, and WTDBG (Table 2). The main reason could be the embedded base–error correction step in CANU that differentiates it from other assemblers. While WTDBG was the fastest and easiest assembler tested, the overall consensus of its final contigs was relatively low (<50% of BUSCO completion). However, after polishing the contigs with Illumina reads (>94% BUSCO completion; verified with a developer) and/or correcting errors with Racon (>83% BUSCO completion), the overall final consensus was good enough to be used in comparisons with other assemblers’ outcomes. While all contigs generated from these assemblers required further polishing with long/short reads to increase the overall accuracy, extra cautious steps were required for MMR, SMD, and WTDBG.
Postprocessing Assembly Performance
The second stage of an assembly is to assemble the conserved regions of the genome to reduce the complexity of de novo assembly for the nonconserved portions. While the quality of input assemblies is expected to directly affect that of the final merged assembly, we have not explored different input qualities in this paper. QMG was employed as a single reconciliation tool to evaluate postprocessing assembly performance because it allows users to merge an assembly obtained from PacBio reads alone or with another assembly based on second-generation reads.(24)
QMG was run with the default parameters, and BUSCO scores were used to gather extensive assembly statistics and gene content completion.(25) While merging multiple individual assemblies substantially improved both the contig size and number (with general improvements to contiguity) for the majority of species, the BUSCO values maintained similar quality statistics with the single best outcomes in preprocessing. In general, as the number of inputs increased, the contiguity improved, thus resulting in fewer but longer contigs. Even merging the two best outcomes from CANU and Flye showed improvements (data not shown). Although we did not investigate further misassemblies, Alhakami et al. (2017) proposed that more inputs for postprocessing assembly could decrease misassemblies and improve contiguity.(17) Details on the statistics and BUSCO from QMG can be found in Figures 2 and 3.
Discussion
ARTICLE SECTIONS
Jump To
NGS technologies and sequence data analysis (including de novo assembly) have radically transformed the field of plant genomics in recent years. Substantial advancements in LRSs and bioinformatics have also provided the necessary framework to systematically analyze data.(3) However, determining the most effective way to sequence and assemble a large, complex plant genome (>1 Gb) among the increasingly varied sequencing and assembly approaches remains difficult. In particular, the existence of long-read assemblers mainly focused on model species (e.g., humans and bacterial genomes) have made it more difficult to gauge their capability and efficiency in handling plant genome data, which can be larger and more repetitive.(3,18)
Over the last decade, DBG assembly coupled with short reads of NGSs has been the method of choice to sequence and assemble plant and animal genomes. However, OLC assemblers, such as PacBio and Nanopore, are well-suited to de novo assembly from long reads.(3,6,10,26) Our PacBio RSII assemblies using five different assemblers (preprocessing) have provided varied results, indicating that different algorithms and/or pipelines can affect assembly quality. In general, estimating the assembly quality requires several statistical evaluations: (1) overall assembly size (match the estimated genome size), (2) measures of assembly contiguity (metrics of N50 from contig numbers, longest contigs, and mean contig size), (3) assembly likelihood scores (calculated by aligning reads against each candidate assembly),(27) (4) accuracy of assembly (aligning the contigs to existing physical maps if available), and (5) completeness of the genome assembly (BUSCO).(2,28) While we have not conducted all statistical evaluations, our outcomes based on the five assemblers studied have achieved unprecedented contiguity compared to that of NGSs, and three assemblers (MMR, SMD, and WTDBG) showed satisfactory results with proper correction with Racon. However, it should be noted that PacBio sequencing and analysis comes with higher sequencing costs, error rates, computational power, and computing time compared to Illumina sequencing and analysis. Thus, obtaining a minimum 50X coverage is recommended to produce a high-quality diploid genome (50% more coverage for polyploidy).(3,18) Additionally, extra Illumina reads polished using Pilon (or equivalent tools) would help minimize any residual and/or artefact errors of PacBio. Despite the genome size, if Illumina reads are available for polishing utilizing the two lightweight tools of MMR and WTDBG, this approach would be good for a novice user, as it does not have a steep learning curve and/or heavy computational requirements. However, heavyweight tools, such as CANU and Flye, should be the first choice for an assembler when looking to achieve accurate assemblies. While we did not succeed in getting Falcon to work in our high-performance cluster because it is designed for Sun Grid Engine and not PBSpro, several papers have already proven its capability and efficiency.(18,29)
A comparison of the results between the current study and the previous works has provided another valuable point to consider for selecting a proper assembler (Tables 1 and 3). However, these tables should be interpreted cautiously because the previous works were assembled with early versions of the algorithm and/or pipeline. In the case of Arabis indica, for example, the results from two different versions (ver. 1.3 for Du et al., 2017 and ver. 1.7 for current study) show inconsistent assembly outcomes, although the same default parameters are set in CANU. These might be the bubble issues to avoid false breaks (e.g., repeats) and potential improvement of the autoset error rate. This point is already clarified in CANU’s GitHub (issue numbers #245 and #852 on https://github.com/marbl/canu/releases). Although it would be problematic to compare the assembly outcomes of A. indica with two different CANU versions, it would be a great challenge for software and algorithm developers to improve the high error rate of long-read assemblers. Because there is no single definitive assembler that guarantees to deliver the best result for a given dataset, we highly recommend selecting the best assembly outcome after comparing the latest versions of a minimum of two different assemblers (e.g., CANU and Flye). It is highly likely that different assembly pipelines could generate different results even for the same dataset. The advantages and disadvantages of the five tested assemblers have been summarized to provide a useful guideline for selecting a proper pipeline according to five criteria (memory intensity, running time, BUSCO completion, ease of use, and program update) (Figure 4). According to our experience and tested dataset, if the coverage is more than 70X and computational resources are not limited, then selecting CANU and Flye is the optimal choice regardless of repeat content and ploidy issues in plant genome assembly. In addition, particularly in the case of CANU setting, a stringent length filtering option (minOverlapLength >3000 or even higher) after removing all non-nuclear genome data (i.e. chloroplast and mitochondrial DNA) results in more correct assemblies.
Figure 4
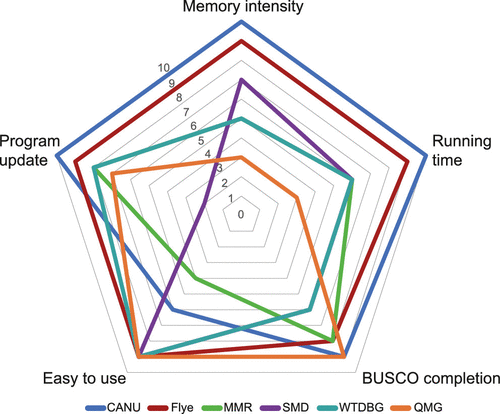
Figure 4. Asset pentagons for five different genome assemblers. Comparison of assembly performance and recommendation for all five assemblies.
Table 3. Comparison of Assembly Results between the Current Study and Previous Worksa
| A. alpina | O. indica | D. zibethinus | C. quinoa | Z. mays | ||
|---|---|---|---|---|---|---|
| Current Study | ||||||
| CANU | TNCb | 797 | 2156 | 6894 | 3521 | 17,653 |
| TACSc | 331 | 363 | 707 | 1265 | 2093 | |
| N50d | 1.6 | 0.3 | 0.4 | 1.3 | 0.2 | |
| Flye | TNCb | 6151 | 1937 | 13,238 | 7251 | 19,038 |
| TACSc | 244 | 358 | 584 | 1011 | 2118 | |
| N50d | 0.1 | 0.4 | 0.1 | 0.3 | 0.2 | |
| MMR | TNCb | 5004 | 2368 | 16,740 | 28,009 | 30,156 |
| TACSc | 379 | 385 | 810 | 1683 | 1249 | |
| N50d | 0.2 | 0.3 | 0.1 | 0.1 | 0.1 | |
| SMD | TNCb | 952 | 2664 | 4670 | 8865 | 26,875 |
| TACSc | 327 | 378 | 711 | 999 | 2151 | |
| N50d | 0.1 | 0.2 | 0.5 | 0.2 | 0.2 | |
| WTDBG | TNCb | 2522 | 2186 | 7953 | 6272 | 27,328 |
| TACSc | 329 | 383 | 823 | 1216 | 2198 | |
| N50d | 0.6 | 0.4 | 0.6 | 0.6 | 0.2 | |
| QMG | TNCb | 1220 | 1429 | 7148 | 1959 | 14,103 |
| TACSc | 550 | 464 | 1462 | 1389 | 2198 | |
| N50d | 1.3 | 0.7 | 0.7 | 1.9 | 0.4 | |
| Previous Works | ||||||
| CANU | TNCb | 1226 | ||||
| TACSc | 405 | |||||
| N50d | 0.9 | |||||
| Falcon | TNCb | |||||
| TACSc | 328 | |||||
| N50d | 0.8 | |||||
| PBcRe | TNCb | 3822/2045 | ||||
| TACSc | 347 | 471/436 | ||||
| N50d | 0.9 | 0.4/1.1 | ||||
| PBcR-MHAP | TNCb | 2958 | ||||
| TACSc | 2104 | |||||
| N50d | 1.2 | |||||
| Celera Assembler | TNCb | 4232 | ||||
| TACSc | 1325 | |||||
| N50d | 1.7 | |||||
a Focused on contigs, not scaffolds. Only scaffold information was available for D. zibethinus. Empty cells: Data not available.
b Total number of contigs.
c Total assembled contig size (Mb).
d Nb50 of contigs (Mb).
e PBcR; PacBio corrected reads pipeline.
Highly contiguous and accurate plant genome assemblies have been shown to generate in a de novo manner solely using PacBio data, but the final assembly is still not entirely finished with results of one solid sequence per chromosome.(3) Given the practical challenges of a de novo assembly, the idea of reconciliation (postprocessing) is very appealing because it can allow the merging of all assemblies (generated from multiple assemblies) to obtain a high-quality consensus assembly.(17) Regarding the outcomes, the expectation is that the quality of the merged assembly should be at least as good as the best assembly in the input because one should expect the consensus assembly to inherit good qualities from the given inputs. However, it is difficult to produce a merged assembly that is consistently better than (or at least as good as) the given input assemblies. There were two cases in which the consensus assembly (Oryza sativa indica and Chenopodium quinoa) was better than the given inputs, but the merged assembly was relatively good for the majority of the inputs, as was demonstrated in previous work.(17) While we do not screen any potential chimeric assemblies from postprocessing, users should be mindful about introducing chimeric/fusion assemblies from prior merging steps into later steps, particularly for polyploid genomes.
Although the current study is limited in scope and use of data, it allows some suggestions to improve future plant and crop genome assemblies: (1) the major availability of the PacBio data is RS II at the moment because we are unable to access a substantial amount of PacBio SEQUEL data to employ various plant species from the public repository. PacBio SEQUEL II has improved over the last 12 months, and this sequencing platform can produce up to 15 times more data per cell (?150 Gb) with higher accuracy in longer reads (reads could be ABI Sanger quality up to 40 kb) and reduced sequencing costs. (2) For analytical tools and assembly algorithms, more sophisticated and computationally efficient assemblers have continually been updated; these include CANU (ver. 2.0),(20) Flye (ver. 2.7.1),(21) and WTDBG (ver. 2.2).(30) It is highly recommended to use the latest versions because these assemblers can achieve more contiguity and accuracy in genome assemblies by fixing many known issues and bugs from previous versions. (3) In eukaryotic contigs, the terminal regions could be scanned using a tandem repeat finder(31) for the presence of telomeres that might be related to the peak computational memory in the form of maximum resident set size and CPU times. While our attempt does not compare the comprehensive tandem repeat, error correction, and polishing stages, a cautious approach should be taken because redundant base pairs in the overlapping terminal regions of fragmented contigs lead to unresolved errors, even after several rounds of consensus polishing. (4) Our attempt highlights how best to use limited genomic resources for effectively evaluating the de novo genome assemblies and performances of plant and crop species for novice users (nonexpert in bioinformatics). Furthermore, it provides a minimal advisable requirement of RAM and CPU cores by comparing assembly metrics and BUSCO completion. However, when completed high-quality genome references are available, the most comprehensive genome assembly comparison could be achievable. For example, (i) single-nucleotide variations (SNVs) and indels and structural variations (SVs) could be useful to evaluate assembly correctness and provide a relative measure of assembly errors and (ii) dot plots could be informative to visualize the genomic variations and rearrangements. Despite not having a chance to test new HiFi data, the updated assemblers, and all criteria, according to our outcomes, selecting any of the three de novo genome assemblers of CANU, Flye, and MMR would allow novice users to acquire suitable results in plant genome assemblies. Utilizing QMG as a postprocessing assembly step is a good strategy depending on the outcomes of the preprocessing assembly.
Impressive strides have been made in the production of plant genome assemblies, thanks to the availability of high-throughput LRSs and NGS data and improved assembly tools/algorithms.(3) For researchers, selecting the best sequencing platform and analytical approach for genome assembly remains challenging, as each option has pros and cons. Nevertheless, continued advances in both sequencing and bioinformatic technologies increase the likelihood of delivering accurate, contiguous, and eventually entire chromosome sequences at low costs. We hope that the comparison of the long-read assemblers we have tested will aid and encourage researchers to spend less time on genome assembly and focus more on exploring the biology of genomes to achieve their research goals.
Supporting Information
ARTICLE SECTIONS
Jump To
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jafc.0c01647.
-
Examples of the executed codes CANU, Flye, MMR, SMD, WTDBG, and QuickMerge (PDF)
-
pdf
- jf0c01647_si_001.pdf (59.23 kb)
Comparative Evaluation of Genome Assemblers from Long-Read Sequencing for Plants and Crops
29views
0shares
0downloads
Skip to figsharenavigation
Supplementary Informat
ion
A comparative evaluation of genome assemblers from long
-
read sequencing
for plants and crops
Hyungtaek Jung
1
,
Min
-
Seung Jeon
2
,
Matthew Hodgett
3
,
Peter Waterhouse
1
, S
eong
-
il Eyun
2
*
1
Centre for Agriculture and Biocommodities
, Queensland University of Technology,
Brisbane,
QLD 4001,
Australia
2
Department of Life Science, Chung
-
Ang University, Seoul
06974
, Korea
3
Information Technology Services, Queensland University of Technology, Brisbane,
QLD
4001,
Australia
*
Corresponding Authors:
Hyungtaek Jung (h7.jung@qut.edu.au)
and
Seong
-
i
l Eyun
(
eyun@cau.ac.kr
)
Supplementary Script S1.
E
xamples of the executed co
des.
C
ANU
canu
\
-
p PRJCA
-
d /home/CANU_PRJCA000313
\
genomeSize=0.5g
\
-
pacbio
-
raw
/home/PRJCA000313_Indica/PacB_Indica.fastq
\
maxMemory=140
\
minMemory=
4
\
correctedErrorRate=0.039
\
useGrid=true
\
gridOptionsJobName=PROsi
\
"gridOptions=
-
V
-
l walltime=15
0
:00:00
-
W umask=0007"
\
minReadLength=1000
\
minOverlapLength=1000
\
Flye
flye
--
pacbio
-
raw /home/PRJCA000313_Indica/PacB_Indica.fasta
--
genome
-
size 500m
--
out
-
dir Flye_Ind
--
threads 14
MMR
minimap
-
Sw5
-
L100
-
m0
\
-
t 12
\
/home/PRJCA000313_Indica/PacB_Indica.fastq
\
/home/PRJCA000313_Indica/P
acB_Indica.fastq
\
| gzip
-
1 > PR313_RCog/P313RC.paf.gz
miniasm
-
Rc2
-
f /home/PRJCA000313_Indica/PacB_Indica.fastq
\
PR313_RCog/P313RC.paf.gz
\
> PR313_RCog/P313RC.gfa
awk '/^S/{print ">"$2"
\
n"$3}' PR313_RCog/P313RC.gfa | fold >
PR313_RCog/P313RC.gfa.fasta
# Correction 1
minimap
-
t 12 PR313_RCog/P313RC.gfa.fasta
\
/home/PRJCA000313_Indica/PacB_Indica.fastq
\
> PR313_RCog/P313RC.gfa1.paf
/work/racon/bin/racon
--
bq
-
1
-
t 10
\
ShareDownload
figshare
This project was supported by a Chung-Ang University Research Grant in 2017 to SE and an Australian Research Council (ARC) Laureate Fellowship (LF160100155) to PW.
The authors declare no competing financial interest.
Terms & Conditions
Electronic Supporting Information files are available without a subscription to ACS Web Editions. The American Chemical Society holds a copyright ownership interest in any copyrightable Supporting Information. Files available from the ACS website may be downloaded for personal use only. Users are not otherwise permitted to reproduce, republish, redistribute, or sell any Supporting Information from the ACS website, either in whole or in part, in either machine-readable form or any other form without permission from the American Chemical Society. For permission to reproduce, republish and redistribute this material, requesters must process their own requests via the RightsLink permission system. Information about how to use the RightsLink permission system can be found at http://pubs.acs.org/page/copyright/permissions.html.
Acknowledgments
ARTICLE SECTIONS
Jump To
The authors are grateful to their colleagues/collaborators, the field/technical specialists of each company, and the three anonymous reviewers for their valuable comments. The authors are especially grateful to Michal Lorenc, QUT High-Performance Computing and Research Support, and the eResearch team for their technical assistance.
| Abbreviations | |
| DBGe | Bruijn Graph |
| HPC | high-performance computer |
| GAGE | genome assembly gold-standard evaluation |
| LRS | long-read sequencing |
| MMR | Miniasm/Minimap/Racon |
| NGS | next-generation sequencing |
| OLC | overlap-layout-consensus |
| PacBio | Pacific Biosciences |
| SMD | SMARTdenovo |
| QMG | Quickmerge |